Description
Information
Ligation Sequencing Kit
This kit is recommended for users who:
want to optimise their sequencing experiment for throughput
require control over read length
would like to utilise upstream processes such as size selection or whole genome amplification.
The Ligation Sequencing Kit features
| Feature | Property |
| Preparation time | 60 minutes |
| Input requirement | 1000 ng of double-stranded DNA |
| PCR Required | No |
| Fragmentation | Optional; recommended for inputs 100-500 ng |
| Read length | Equal to fragment length |
| Read type produced | 1D |
| Typical throughput* | ••• |
| Multiplexing options |
PCR Barcoding Expansion 1-12 and 1-96 Flow Cell Wash Kit |
| Pack size | 6 reactions |
| Stability | Shipped at 2–8°C Long-term storage -20°C Oxford Nanopore Technologies deem the useful life of the product to be 3 months from receipt by the customer. |
The Ligation Sequencing Kit offers a flexible method of preparing sequencing libraries from dsDNA (e.g. gDNA, cDNA or amplicons). The library preparation method is straightforward: DNA ends are repaired and dA-tailed using the NEBNext End Repair/dA-tailing module, and then sequencing adapters, supplied in the kit, are ligated onto the prepared ends.
The kit is optimised to generate maximum throughput and read length. For highest data yields, we recommend starting with 100-200 fmol of pure input DNA. Starting with lower amounts of input material, or impure samples, can affect library preparation efficiency and can reduce sequencing throughput.
The kit contains an updated sequencing adapter (Adapter Mix F, or AMX-F), which is designed to turn over significantly less fuel during a sequencing run compared to previous kit versions. This results in better maintenance of enzyme speed and higher sequencing output.
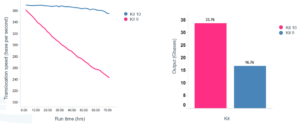
translocation
Users can either start with 1 μg of gDNA, quantified using the Qubit fluorometer, or 100-200 fmol of shorter-fragment input such as amplicons or cDNA. If your experiment requires long reads, it is recommended to start with full-length gDNA, and fragmentation/shearing is neither advised nor required. Please note: if long reads are required, then long fragments must be present in the starting material. To determine purity, we suggest using the Nanodrop to measure the A260/280 and A260/230 ratios and we recommend that the sample should meet the following criteria:
- A260/280 = 1.8
- A260/230 = 2.0-2.2
The Ligation Sequencing Kit is compatible with upstream processes such as target enrichment by sequence capture, whole genome amplification (for applications where under 1 ng of sample is available) and size selection (for enrichment of specified fragment lengths, using the BluePippin, for example). When size selecting, we recommend increasing the amount of input used, as size selection can be a wasteful process.
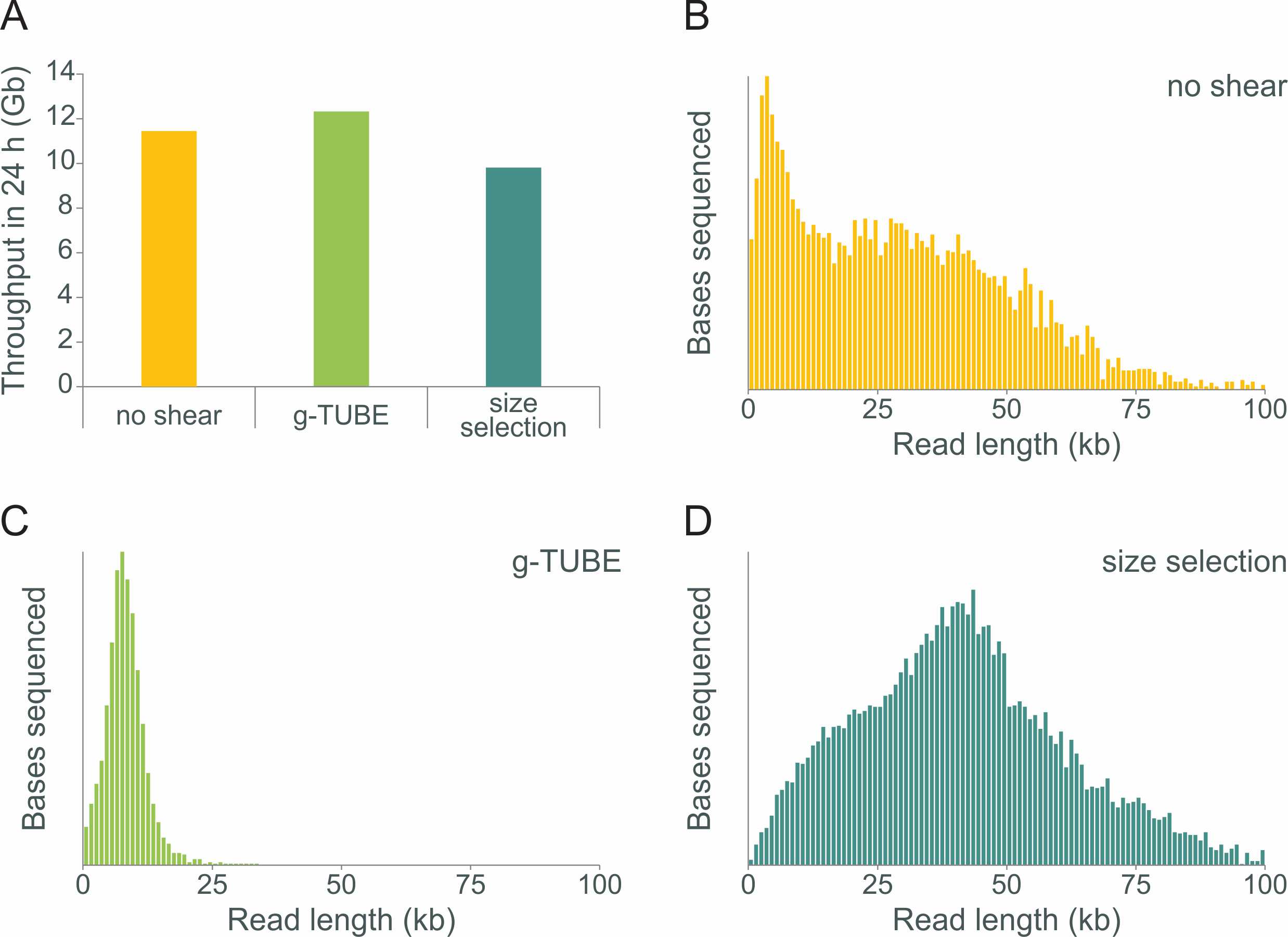
The relative sequencing yields (A) and typical read-length histograms (B, C and D) observed when preparing E.coli libraries using the Ligation Sequencing Kit without or with g-TUBE fragmentation or with size selection (without fragmentation).
PCR- and WGA-free workflows remove amplification bias and retain base modification information, which can be analysed using tools developed in the Nanopore Community.
Further considerations:
Starting with lower amounts of input material, or impure samples, can affect library preparation efficiency and can reduce sequencing throughput. PCR can be used to generate more input material in cases where sample amount is limiting. Also, where sample purity is compromised and library preparation efficiency may be reduced, PCR will select for molecules which have been successfully adapted, and generate more of them. The original impurity is lost or diluted. It is often observed that a sample which produces poor sequencing results can be improved by inclusion of PCR. However, if PCR is likely to be the main method of library preparation, we recommend the PCR Sequencing Kit, or Rapid PCR Sequencing Kit.
While any dsDNA can be used as a template for the Ligation Sequencing Kit, users planning to regularly sequence amplicons or cDNA, should consider specific kits with dedicated protocols which simplify the laboratory workflow and produce superior results. For amplicons, we recommend using our four-primer PCR protocol along with the PCR Sequencing Kit (or PCR Barcoding Kit for multiplexing amplicons), and for cDNA we recommend the PCR-cDNA Sequencing Kit or the Direct cDNA Sequencing Kit.
Shipping and logistics
Flow cells and kits are shipped together at 2-8°C. Upon receipt, place the product in the right long-term storage location.
Products are shipped to customers within the USA and EU Monday to Thursday. Shipments to Canada, Norway, Korea and Japan are expedited Monday to Wednesday; with Australia and New Zealand leaving our warehouses on a Friday. Shipments to the rest of the world are made on Mondays to allow the full working week for packages to arrive.
The delivery charges are calculated when a quote is raised or during checkout. Once an order is made, the delivery ID and delivery information can be tracked in the Store.
Workflow
The Ligation Sequencing Kit offers a flexible method of preparing sequencing libraries from dsDNA (e.g. gDNA, cDNA or amplicons). The library preparation method is straightforward: DNA ends are FFPE repaired and end-prepped/dA-tailed using the NEBNext End Repair/dA-tailing module, and then sequencing adapters, supplied in the kit, are ligated onto the prepared ends.
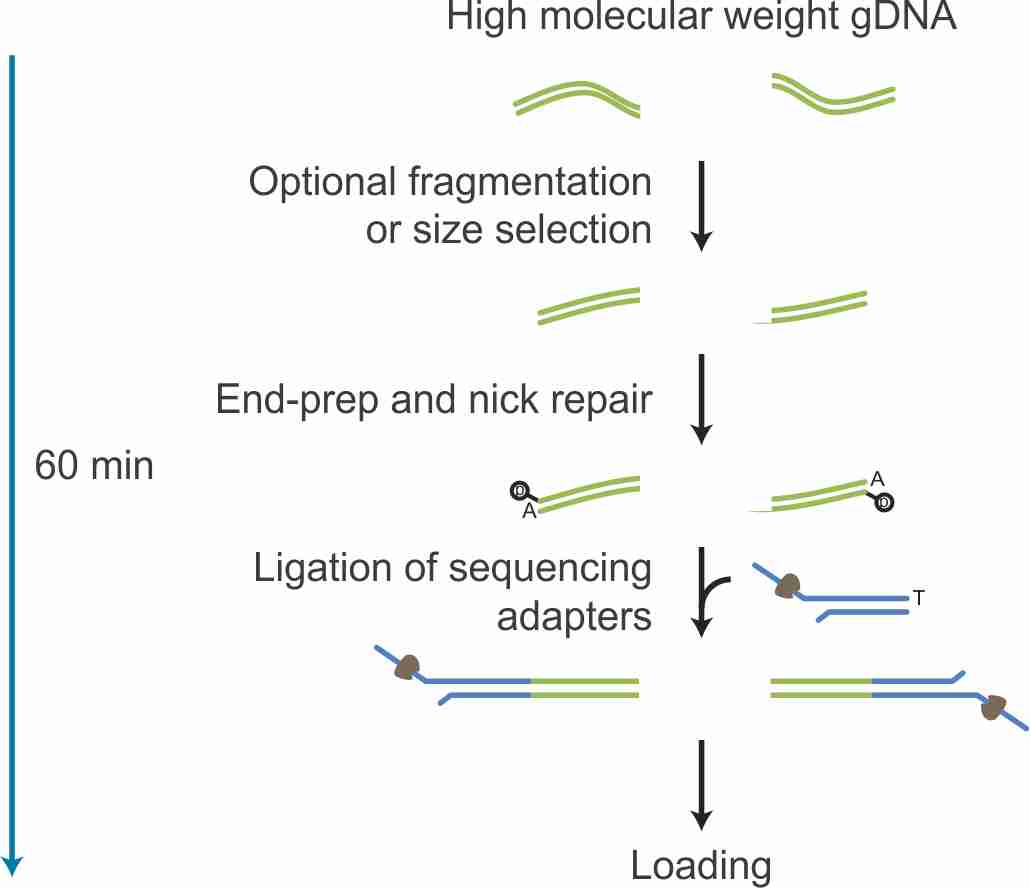
ligation_sequencing_kit_workflow
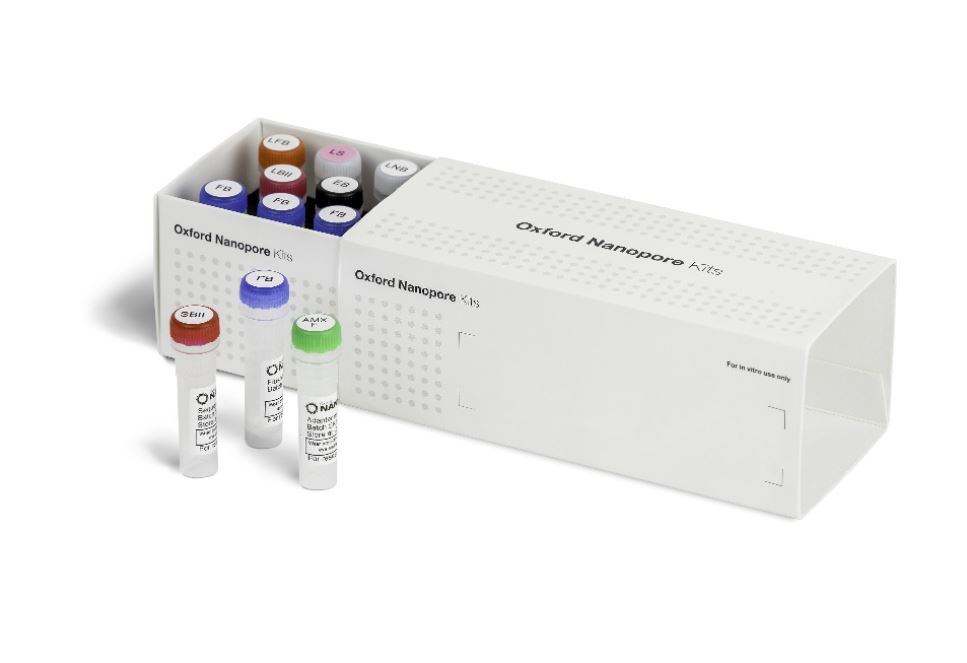
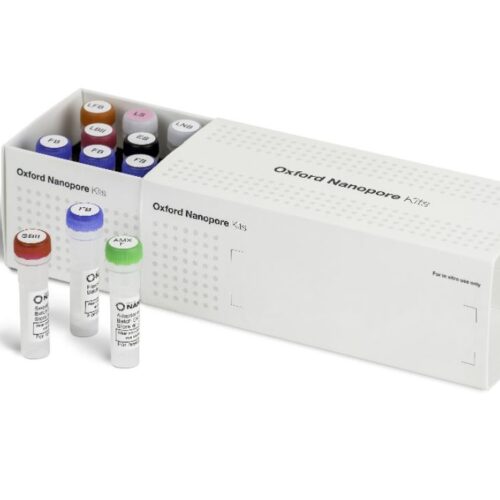
What’s in the box
The Ligation Sequencing Kit contains sufficient reagents to generate six sequencing libraries.
ligation-fragmentation_kit_contents
Compatibility
The Ligation Sequencing Kit can be used together with:
Kits
- PCR Barcoding Expansion 1-12 (EXP-PBC001)
- PCR Barcoding Expansion 1-96 (EXP-PBC096)
- PCR Expansion (EXP-PCA001)
- Sequencing Auxiliary Vials (EXP-AUX110)
- Flow Cell Wash Kit (EXP-WSH004)
- Native Barcoding kit upgrades coming soon
Flow cells
- FLO-MINSP6
- FLO-MIN106D
- FLO-MIN111
- FLO-PRO002
- FLO-FLG001
Devices