Description
Information
This kit is recommended for users who:
wish to multiplex samples to reduce price per sample
have a limited amount of input material
want to optimise their sequencing experiment for throughput
would like to identify and quantify full-length transcripts
are interested in differential gene expression or differential transcript usage
want to characterise and quantify isoforms, splice variants and fusion transcripts
The PCR-cDNA Barcoding Kit features:
| Features | PCR-cDNA Barcoding Kit | |
| Preparation time | 165 mins | |
| Input requirement | 1 ng poly-A+ RNA, or 50 ng total RNA | |
| RT Required | Yes | |
| PCR required | Yes | |
| Read length | Enriched for full-length cDNA during PCR | |
| Read type produced | 1D | |
| Typical throughput | ••• | |
| Typical number of reads (on FLO-MIN106 at an average read length of 1 kb) | 7-12+ million full-length mapped reads per flow cell on MinION/GridION; 50+ million mapped reads per flow cell on PromethION | |
| Associated protocols | PCR-cDNA Barcoding Kit
Flow Cell Wash Kit |
|
| Pack size | 6 reactions | |
| Storage | Shipped at -20° C
Long term storage -20° C |
|
Description:
Barcoding or multiplexing is useful when the amount of data required per sample is less than the total amount of data that can be generated from a single flow cell: it allows a user to pool multiple samples and sequence them together, making more efficient use of the flow cell.
The PCR-cDNA Barcoding Kit is used to prepare cDNA for nanopore sequencing for up to 12 samples, from an input of as low as 1 ng poly-A+ RNA. Users who do not have poly-A+ enriched RNA can use 50 ng of total RNA but additional optimisation may be required.
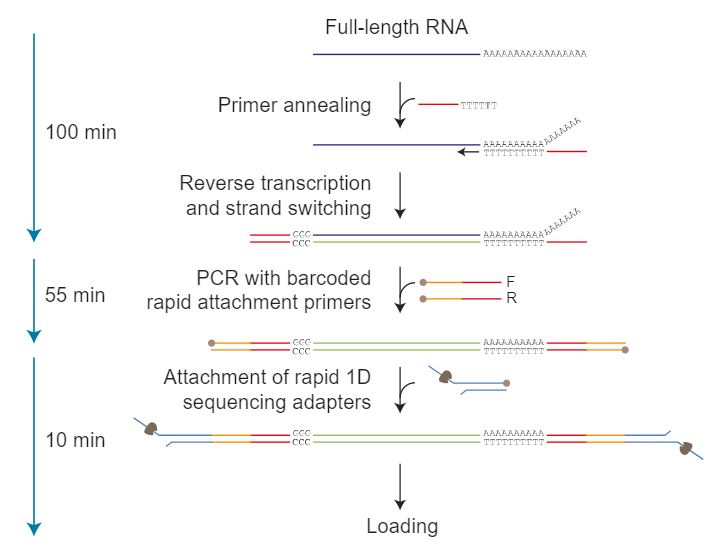
Taking full-length poly-A+ RNA, complementary strand synthesis and strand switching are performed using kit-supplied oligonucleotides. The kit contains 12 primer pairs which are used to generate and then amplify double-stranded cDNA by PCR amplification: each primer pair contains a barcode and 5’ tag which facilitates the ligase-free attachment of Rapid Sequencing Adapters. Amplified and barcoded samples are then pooled together, and Rapid Sequencing Adapters are added to the pooled mix.
Shipping and logistics:
Flow cells and kits are shipped together in a temperature-controlled shipping box with gel ice packs. Ice packs are stored at 2-8° C, and -20° C ice packs are added to the top of the box (but not in direct contact with product) to maintain the integrity of the shipment.
Products are shipped to customers within the USA and EU Monday to Thursday. Shipments to Canada, Norway, Korea and Japan are expedited Monday to Wednesday; with Australia and New Zealand leaving our warehouses on a Friday. Shipments to the rest of the world are made on Mondays to allow the full working week for packages to arrive.
The delivery charges are calculated when a quote is raised or during checkout. Once an order is made, the delivery ID and delivery information can be tracked in the Store.
Oxford Nanopore Technologies deem the useful life of the product to be 3 months from receipt by the customer.
Workflow
Taking full-length poly-A+ RNA, complementary strand synthesis and strand switching are performed using kit-supplied oligonucleotides. The kit contains 12 primer pairs which are used to generate and then amplify double-stranded cDNA by PCR amplification: each primer pair contains a barcode and 5’ tag which facilitates the ligase-free attachment of Rapid Sequencing Adapters. Amplified and barcoded samples are then pooled together, and Rapid Sequencing Adapters are added to the pooled mix.
What’s in the box
The 16S Barcoding Kit 1-24 contains 24 unique barcodes and sufficient reagents to generate six sequencing libraries.
The Flow Cell Priming Kit Expansion Pack is also supplied.
Data analysis
Data from a PCR-cDNA sequencing experiment can be analysed using a number of bioinformatics tools and pipelines:
DESeq2 for differential gene expression profiling
pinfish for annotation of gene transcripts and novel gene isoform discovery
pipeline-transcriptome-de for performing differential transcript usage (DTU) and differential gene expression (DGE) analyses
Oxford Nanopore Technologies offers easy-to-follow bioinformatics tutorials, which take the user step-by-step through each analysis pipeline. These tutorials are available in the Nanopore Community.
What’s in the box
The PCR-cDNA Barcoding Kit contains sufficient reagents to generate six sequencing libraries.
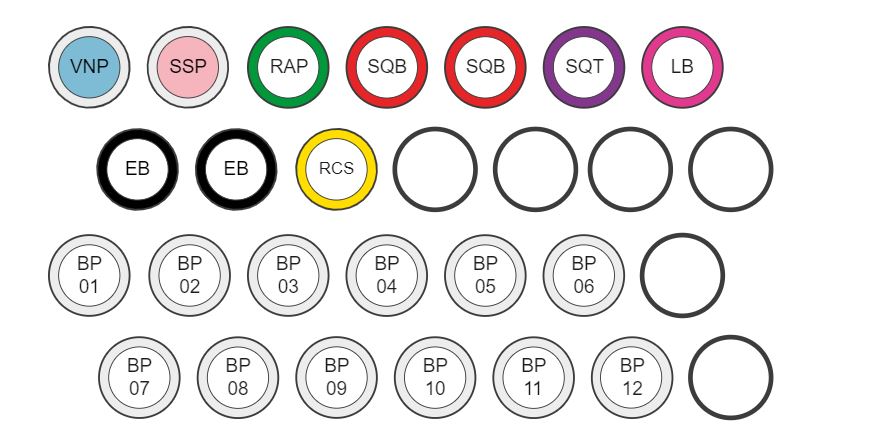
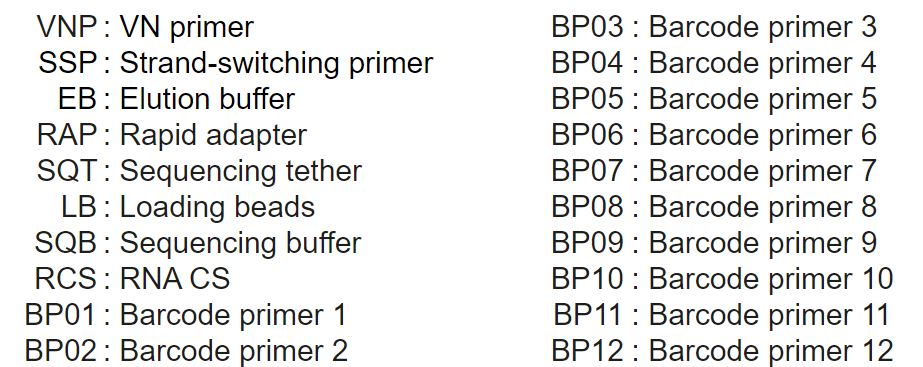
Compatibility
The PCR-cDNA Barcoding Kit can be used together with:
Kits
- Flow Cell Wash Kit (EXP-WSH004)
Flow cells
- FLO-MINSP6
- FLO-MIN106D
- FLO-PRO002
Devices