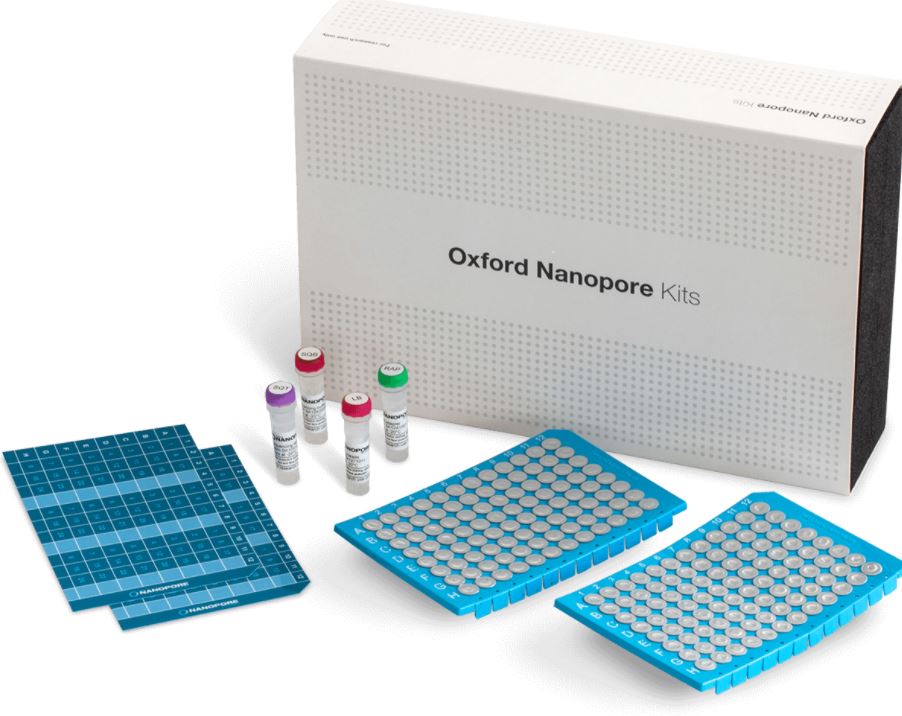
Description
Information
This kit is recommended for users who:
- want to do 16S sequencing
- are interested in genus-level bacterial identification
Barcoding or multiplexing is useful when the amount of data required per sample is less than the total amount of data that can be generated from a single flow cell: it allows a user to pool multiple samples and sequence them together, making more efficient use of the flow cell.
The 16S Barcoding Kit offers a method of amplifying the entire ~1,500 bp 16S rRNA gene from extracted gDNA, followed by library preparation. By narrowing down to a specific region of interest, a user can see all the organisms in the sample without sequencing unnecessary regions of the genome, making the identification quicker and more economical. On the other hand, analysing the full 16s rRNA gene is more informative than analysing a subset of exons.
The 16S Barcoding Kit features:
| Feature | Property |
| Preparation time | 10 minutes + PCR |
| Input requirement | <10 ng gDNA |
| PCR Required | Yes |
| Fragmentation | Not required |
| Read length | Full-length 16S gene (~1.5 kb) |
| Read type produced | 1D |
| Typical throughput* | ••• |
| Associated protocols | 16S Barcoding Kit 1-24 (SQK-16S024) |
| Multiplexing options | 16S Barcoding Kit 1-24
Wash Kit |
| Pack size | 6 reactions |
| Stability | Stable at RT for 7 days
Shipped at 2-8° C Long-term storage -20° C |
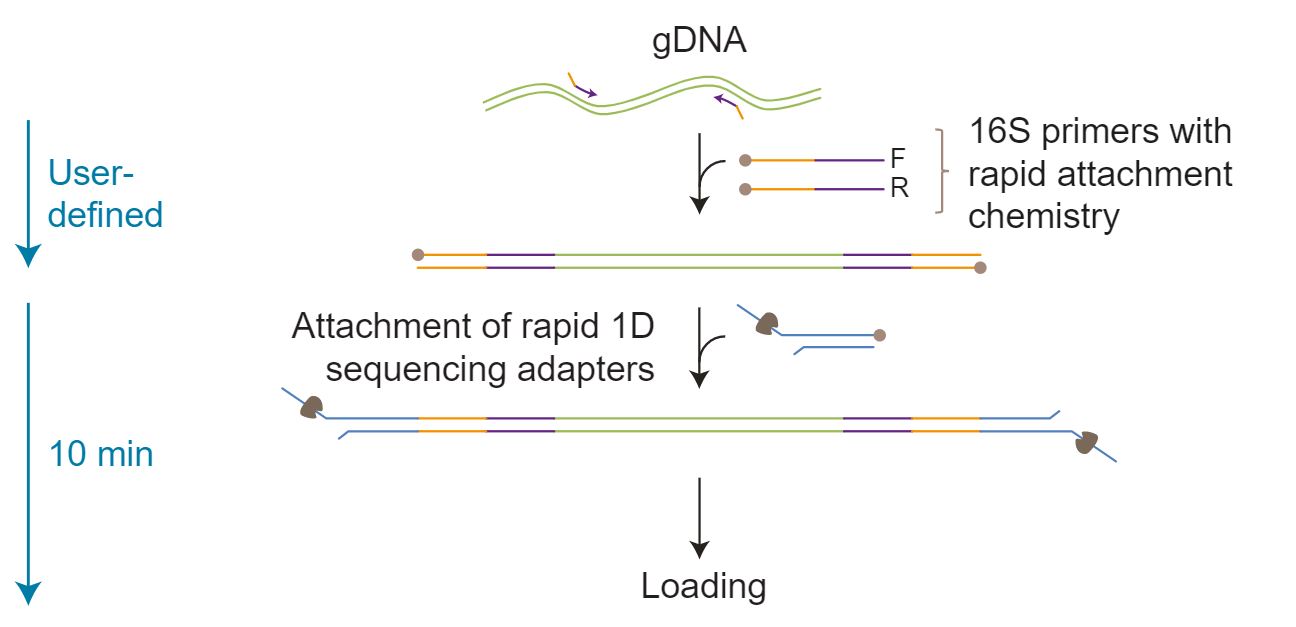
The DNA is amplified by PCR using specific 16S primers (27F and 1492R) that contain 5’ tags which facilitate the ligase-free attachment of Rapid Sequencing Adapters.
The kit is supported by the EPI2ME FASTQ 16S workflow, which can be used to analyse data from the 16S protocol. Deconvolution of barcoded sequencing data is supported by Guppy and EPI2ME which classify the barcode sequence and sort reads into corresponding folders.
Shipping and logistics:
The DNA is amplified by PCR using specific 16S primers (27F and 1492R) that contain 5’ tags which facilitate the ligase-free attachment of Rapid Sequencing Adapters. The kit contains 24x barcoded primer pairs.
The kit is supported by the EPI2ME 16S-BLAST workflow, which can be used to analyse data from the 16S protocol. Deconvolution of barcoded sequencing data is supported by Guppy and EPI2ME, which classify the barcode sequence and sort reads into corresponding folders.
Workflow
The Ultra-Long DNA Sequencing Kit generates sequencing libraries from extracted ultra-high molecular weight gDNA in ~610 minutes. At the heart of the kit is a transposase which simultaneously cleaves template molecules and attaches tags to the cleaved ends. Rapid Sequencing Adapters are then added to the tagged ends, followed by an overnight elution, for example using a Circulomics 5 mm Nanobind disk, to remove free adapter and short DNA fragments.
What’s in the box
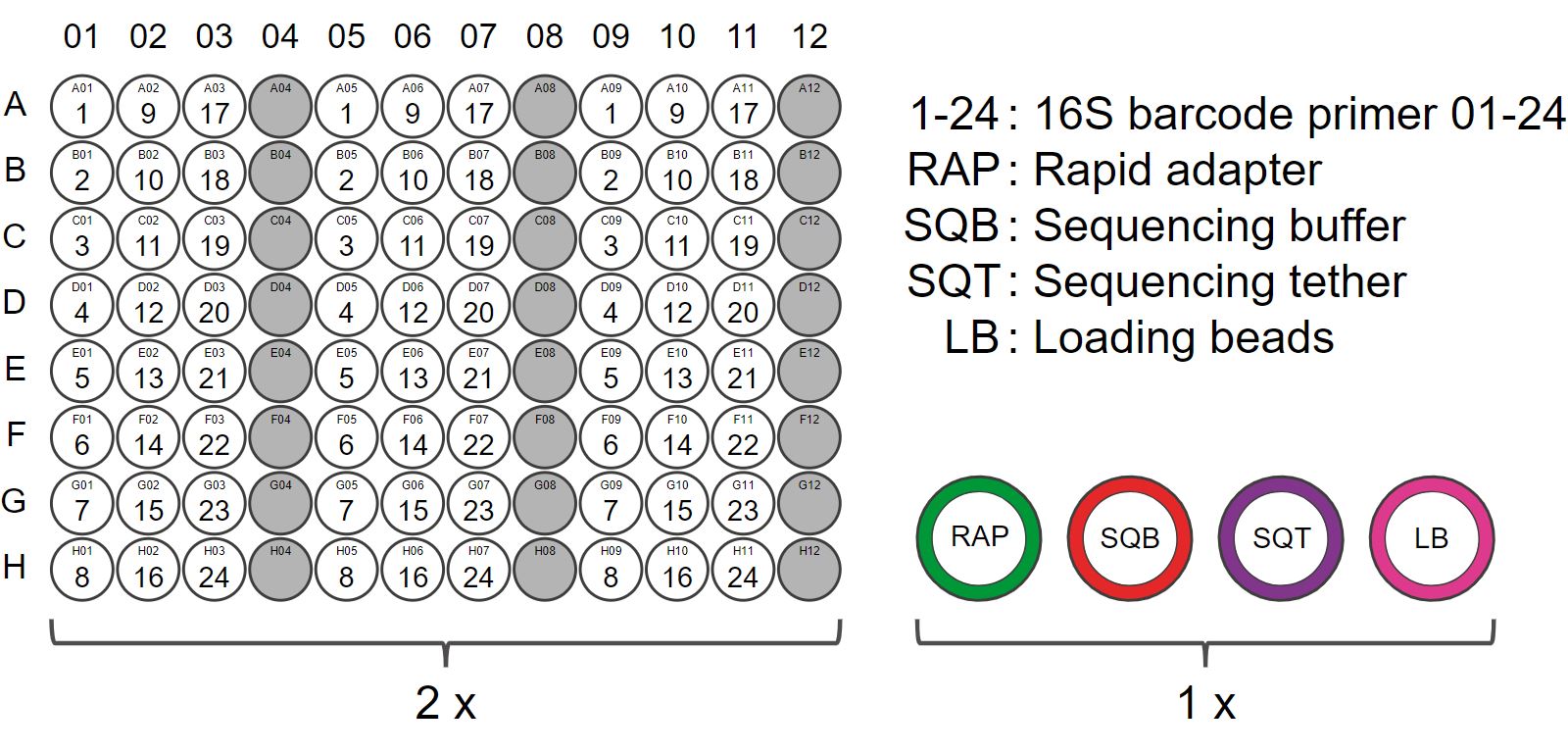
The 16S Barcoding Kit 1-24 contains 24 unique barcodes and sufficient reagents to generate six sequencing libraries.
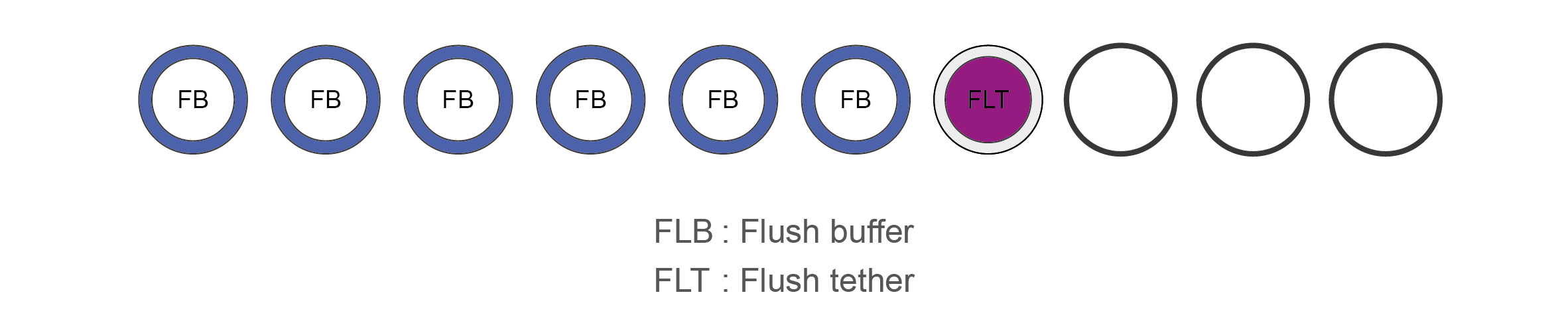
The Flow Cell Priming Kit Expansion Pack is also supplied.
Compatibility
The PCR Sequencing Kit can be used together with:
Kits
- RAP Top-up Kit (EXP-RAP001)
- Flow Cell Wash Kit (EXP-WSH004)
Flow cells
- FLO-MINSP6
- FLO-MIN106D
- FLO-FLG001
Devices